I came across MoLeR a few years ago at a conference in a talk by Krzysztof Maziarz, I liked the fact that it was fast to train, it used graphs rather than text to generate molecules and you could define a scaffold which is typically what you do in almost all projects.
There are loads of generative chemistry programs out there, but I thought MoLeR was particularly well put together and easy to use. For practical applications it was coupled with a molecular swarm optimisation (MSO). Both packages are very good but I had to do a bit of tweaking to get them to play nicely together. Code is available here . Compared to AZ’s Reinvent, I find the MSO + MoLeR combo easier to use, modify and gives less “extreme creative solutions” at least in my hands.
Step one is define is to train a model. This is very straightforward with the instructions on the MoLeR git.
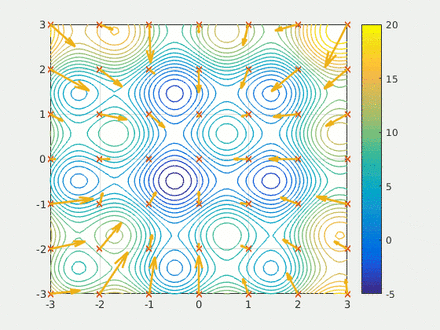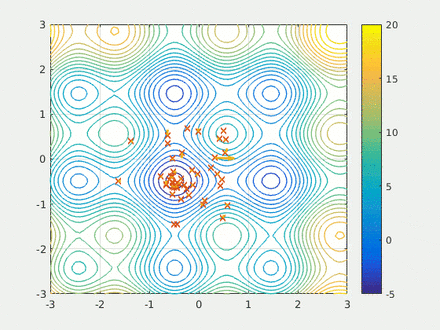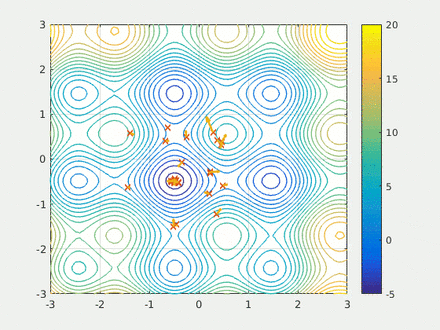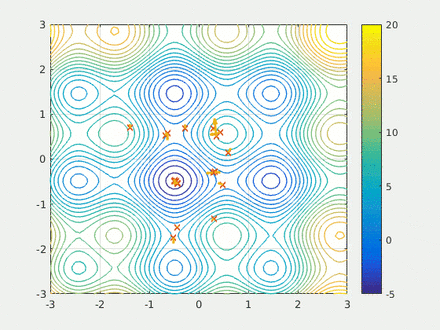
Step two is to define the multidimensional box the swarm(s) will explore. This can be the chemistry used to build the MoLeR model, it can also represents a way of guiding the swarm by selecting a subset of chemistry. Here I encode the smiles with model and return the max and min of the encoding.
python define_max_min_of_model.py <model> <smiles>
Step three define some objectives for the swarm, initial smiles can be anything, here I use benzene but it can also be a swarm of different molecules. The scaffold is enforced and is present in every molecule. The MPO is a simple JSON dictionary. More details are on Robin Winter’s GitHub.
from mso.optimiser import BasePSOptimizer
import numpy as np
from molecule_generation import VaeWrapper
from mso.objectives.swarm_functions import swarm_wt
from mso.objectives.scoring import SwarmScoringFunction, ScoringFunction
from mso.utils import read_model
import time
import os
import sys
init_smiles = "c1ccccc1" # SMILES representation
scaffold = "OC=O"
mwt_desirability = [{"x" : 180, "y" : 0}, {"x":200, "y": 1}, {"x":400, "y": 1.0}, {"x":450, "y": 0.0}]
scoring_functions = [SwarmScoringFunction(func=swarm_wt, name="mwt", desirability = mwt_desirability)]
Step four is running the calculation. Here we have two swarms each with 500 probe molecules, the simple scoring function, the box dimensions, the model and the scaffold. Finally I run the model for 20 epochs and save the results to a csv.
model_dir, x_max, x_min = read_model(sys.argv[1])
with VaeWrapper(model_dir) as model:
opt = BasePSOptimizer.from_query(init_smiles=init_smiles,num_part=500,num_swarms=2,inference_model=model,scoring_functions=scoring_functions, x_max=x_max, x_min=x_min, scaffold=scaffold)
start_time = time.time()
try:
opt.run(20)
timestr = time.strftime("%Y%m%d-%H%M%S")
print("--- %s seconds ---" % (time.time() - start_time))
try:
opt.best_solutions.to_csv("best_solutions_" + timestr + ".csv", index=False)
opt.best_fitness_history.to_csv("best_history_" + timestr + ".csv", index=False)
except:
pass
except:
opt.best_solutions.to_csv("best_solutions_" + "error" + ".csv", index=False)
opt.best_fitness_history.to_csv("best_history_" + "error" + ".csv", index=False)
Leave a comment