Chemprop is my favourite QSAR tool. It’s packed full of features, easy to use, and you can do some fairly sophisticated model building. Here are some notes on running predictions with evidential uncertainty.
Training data
I’ll use the in vogue Biogen dataset from Fang et al. 2023 which can be conveniently accessed via https://polarishub.io . As it’s a toy model I’ll assume the curated data is fine and won’t spend time finding a good data split, tailoring the descriptors, model parameters etc.
pip install git+https://github.com/chemprop/chemprop.git
pip install polaris-lib
pip install -U "ray[data,train,tune,serve]"
We’ll first generate the splits:
import pandas as pd
from sklearn.model_selection import train_test_split
import sys
df = pd.read_csv(sys.argv[1])
# Step 1: Split into training (80%) and remaining data (20%)
train_df, remaining_df = train_test_split(df, test_size=0.2)
# Step 2: Split remaining data into external validation (10%), and calibration (10%)
external_val_df, calibration_df = train_test_split(remaining_df, test_size=0.5)
# reset indices
train_df = train_df.reset_index(drop=True)
external_val_df = external_val_df.reset_index(drop=True)
calibration_df = calibration_df.reset_index(drop=True)
# Write the concatenated DataFrame to a CSV file
train_df.to_csv("training.csv", index=False)
external_val_df.to_csv("ext_val.csv", index=False)
calibration_df.to_csv("cal.csv",index=False)
Model building
chemprop hpopt --data-path training.csv -s "SMILES" --target-columns "AC_LOG SOLUBILITY PH 6.8 (ug/mL)" --task-type regression-evidential --search-parameter-keywords depth ffn_num_layers ffn_hidden_dim message_hidden_dim dropout --hpopt-save-dir hpoptv1 --multi-hot-atom-featurizer-mode ORGANIC -l evidential
Hyperparameter optimization is straightforward but time-consuming; if you’ve got the hardware, it can be sped up with --raytune-use-gpu --raytune-num-cpus 12. I found this very slow to do on an old MacBook Air M1 (>24 hours and still not complete), but Colab came to the rescue, and a free tier TPU instance worked really well (ca. 1 hr). The --multi-hot-atom-featurizer-mode flag with ORGANIC or, more recently, in v2.1.2, RIGR (Resonance Invariant Graph Representation) is more targeted and thus gives better results. At the time of writing, RIGR is a very new addition; so far, in my limited experience, the ORGANIC flag seems to slightly edge out the RIGR flag, but I expect this to be case dependent. From the pre-print, it does sound like the RIGR flag is particularly helpful for small data sets.
With the hyper-parameters identified we are ready to train the model. This can be done with the following command. Number of replicates is analogous to ‘outer loop’ of nested cross validation but at a lower cost, suitable for deep learning applications. Again if the hardware allows the training can be speeded up with --accelerator gpu. Depending on the task it important to specify the appropriate loss function (here evidential), apparently the method choice should pick this up but I am not sure it always does. With evidential regression the --evidential-regularization is model dependent, default is 0.0 and the default recommended by Soleimany et al. (2021) is 0.2
chemprop train --data-path training.csv -s "SMILES" --target-columns "LOG SOLUBILITY PH 6.8 (ug/mL)" --task-type regression-evidential --config-path ./hpoptv1/best_config.toml --multi-hot-atom-featurizer-mode ORGANIC --num-replicates 10 -o model_evidential --aggregation norm --epochs 50 -l evidential --evidential-regularization 0.2
Final step is get the predictions on the external validation set together with the uncertainty estimation which is calibrated with conformal-regression.
chemprop predict -s "SMILES" --test-path ext_val.csv --model-path model_evidential --preds-path preds.csv --multi-hot-atom-featurizer-mode ORGANIC --uncertainty-method evidential-total --calibration-method conformal-regression --cal-path cal.csv
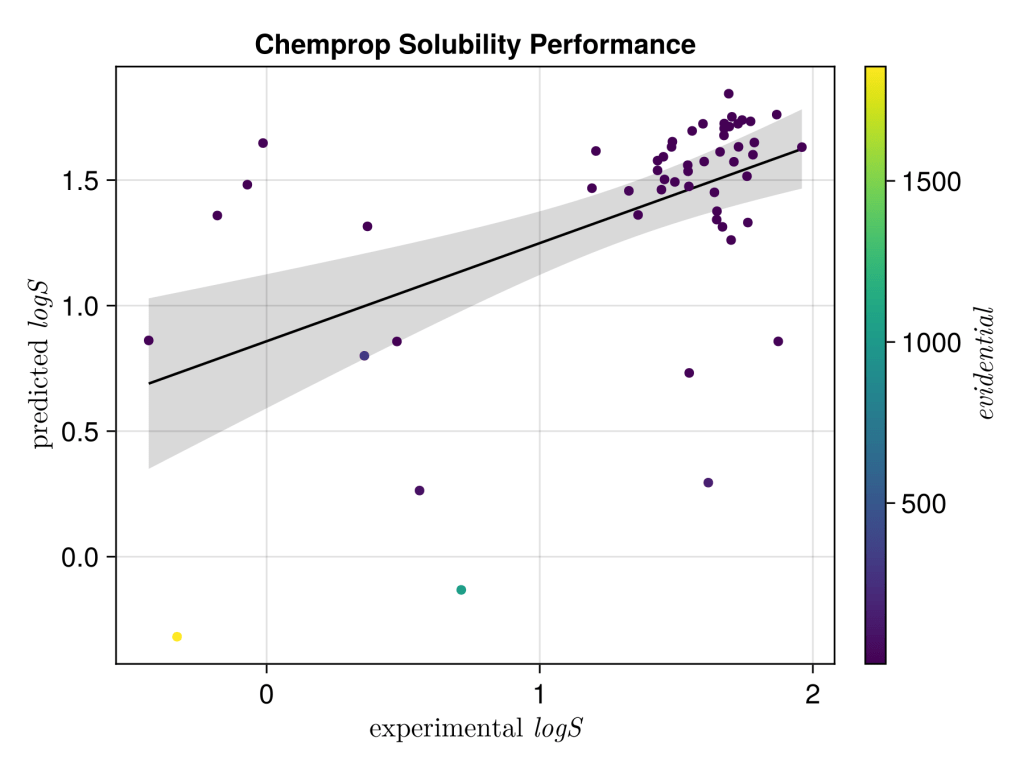
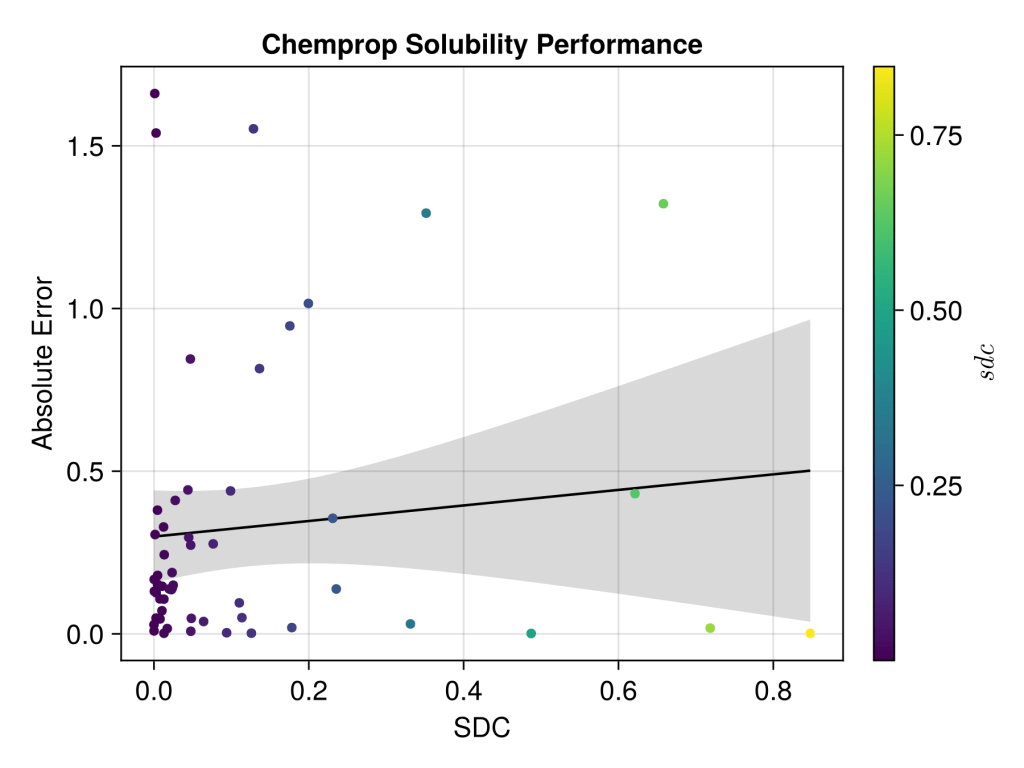
The final results don’t look too impressive, but maybe not unsurprising; it’s a very unsophisticated model, the data range is pretty small with logS, it’s a tricky end point to predict, the experimental values can vary a lot depending on sample, the selected data isn’t well distributed over the experimental range and furthermore the applicability domain calculations suggest test set looks pretty different to train. Uncertainty estimations don’t seem to rescue the model either, or look calibrated… I’ll have to more flattering dataset next time but at least it look realistic. Next step is to see if it can be competitive TabPFN.
Leave a comment